This textbook was written for the clinical research community at Johns Hopkins leveraging the precision medicine analytics platform (PMAP). These notebooks are available in html form on the Precision Medicine portal as well as in computational form in the CAMP-share folder on Crunchr (Crunchr.pm.jh.edu).
XNAT SessionPydicomCurrently, accessing patient DICOMs requires managing several files and loading them.
Using an XNAT Session to programmatically access DICOMs using Jupyter notebook, the user can easily directly load a file instead of having to manage the various files in the file system and loading them one-by-one.
Install/Import XNAT and subsequent complementary packages for viewing and analysis:
Install XNAT using
piponly if not installed already
#pip install xnat
Requirement already satisfied: xnat in /home/idies/miniconda3/lib/python3.8/site-packages (0.4.2) Requirement already satisfied: progressbar2>=3.11.0 in /home/idies/miniconda3/lib/python3.8/site-packages (from xnat) (4.0.0) Requirement already satisfied: six>=1.13.0 in /home/idies/miniconda3/lib/python3.8/site-packages (from xnat) (1.15.0) Requirement already satisfied: isodate>=0.5.4 in /home/idies/miniconda3/lib/python3.8/site-packages (from xnat) (0.6.1) Requirement already satisfied: requests>=2.10.0 in /home/idies/miniconda3/lib/python3.8/site-packages (from xnat) (2.24.0) Requirement already satisfied: click>=7.0.0 in /home/idies/miniconda3/lib/python3.8/site-packages (from xnat) (7.1.2) Requirement already satisfied: sphinx-rtd-theme>=0.1.9 in /home/idies/miniconda3/lib/python3.8/site-packages (from xnat) (0.5.0) Requirement already satisfied: python-utils>=3.0.0 in /home/idies/miniconda3/lib/python3.8/site-packages (from progressbar2>=3.11.0->xnat) (3.3.3) Requirement already satisfied: chardet<4,>=3.0.2 in /home/idies/miniconda3/lib/python3.8/site-packages (from requests>=2.10.0->xnat) (3.0.4) Requirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /home/idies/miniconda3/lib/python3.8/site-packages (from requests>=2.10.0->xnat) (1.25.10) Requirement already satisfied: idna<3,>=2.5 in /home/idies/miniconda3/lib/python3.8/site-packages (from requests>=2.10.0->xnat) (2.10) Requirement already satisfied: certifi>=2017.4.17 in /home/idies/miniconda3/lib/python3.8/site-packages (from requests>=2.10.0->xnat) (2020.6.20) Requirement already satisfied: sphinx in /home/idies/miniconda3/lib/python3.8/site-packages (from sphinx-rtd-theme>=0.1.9->xnat) (3.2.1) Requirement already satisfied: sphinxcontrib-applehelp in /home/idies/miniconda3/lib/python3.8/site-packages (from sphinx->sphinx-rtd-theme>=0.1.9->xnat) (1.0.2) Requirement already satisfied: sphinxcontrib-devhelp in /home/idies/miniconda3/lib/python3.8/site-packages (from sphinx->sphinx-rtd-theme>=0.1.9->xnat) (1.0.2) Requirement already satisfied: alabaster<0.8,>=0.7 in /home/idies/miniconda3/lib/python3.8/site-packages (from sphinx->sphinx-rtd-theme>=0.1.9->xnat) (0.7.12) Requirement already satisfied: babel>=1.3 in /home/idies/miniconda3/lib/python3.8/site-packages (from sphinx->sphinx-rtd-theme>=0.1.9->xnat) (2.8.0) Requirement already satisfied: imagesize in /home/idies/miniconda3/lib/python3.8/site-packages (from sphinx->sphinx-rtd-theme>=0.1.9->xnat) (1.2.0) Requirement already satisfied: setuptools in /home/idies/miniconda3/lib/python3.8/site-packages (from sphinx->sphinx-rtd-theme>=0.1.9->xnat) (49.6.0.post20200814) Requirement already satisfied: Jinja2>=2.3 in /home/idies/miniconda3/lib/python3.8/site-packages (from sphinx->sphinx-rtd-theme>=0.1.9->xnat) (2.11.2) Requirement already satisfied: sphinxcontrib-qthelp in /home/idies/miniconda3/lib/python3.8/site-packages (from sphinx->sphinx-rtd-theme>=0.1.9->xnat) (1.0.3) Requirement already satisfied: sphinxcontrib-serializinghtml in /home/idies/miniconda3/lib/python3.8/site-packages (from sphinx->sphinx-rtd-theme>=0.1.9->xnat) (1.1.4) Requirement already satisfied: docutils>=0.12 in /home/idies/miniconda3/lib/python3.8/site-packages (from sphinx->sphinx-rtd-theme>=0.1.9->xnat) (0.16) Requirement already satisfied: sphinxcontrib-jsmath in /home/idies/miniconda3/lib/python3.8/site-packages (from sphinx->sphinx-rtd-theme>=0.1.9->xnat) (1.0.1) Requirement already satisfied: snowballstemmer>=1.1 in /home/idies/miniconda3/lib/python3.8/site-packages (from sphinx->sphinx-rtd-theme>=0.1.9->xnat) (2.0.0) Requirement already satisfied: packaging in /home/idies/miniconda3/lib/python3.8/site-packages (from sphinx->sphinx-rtd-theme>=0.1.9->xnat) (20.4) Requirement already satisfied: Pygments>=2.0 in /home/idies/miniconda3/lib/python3.8/site-packages (from sphinx->sphinx-rtd-theme>=0.1.9->xnat) (2.7.0) Requirement already satisfied: sphinxcontrib-htmlhelp in /home/idies/miniconda3/lib/python3.8/site-packages (from sphinx->sphinx-rtd-theme>=0.1.9->xnat) (1.0.3) Requirement already satisfied: pytz>=2015.7 in /home/idies/miniconda3/lib/python3.8/site-packages (from babel>=1.3->sphinx->sphinx-rtd-theme>=0.1.9->xnat) (2020.1) Requirement already satisfied: MarkupSafe>=0.23 in /home/idies/miniconda3/lib/python3.8/site-packages (from Jinja2>=2.3->sphinx->sphinx-rtd-theme>=0.1.9->xnat) (1.1.1) Requirement already satisfied: pyparsing>=2.0.2 in /home/idies/miniconda3/lib/python3.8/site-packages (from packaging->sphinx->sphinx-rtd-theme>=0.1.9->xnat) (2.4.7) Note: you may need to restart the kernel to use updated packages.
import xnat
import pydicom
from pydicom.data import get_testdata_files
import matplotlib.pyplot as plt
import numpy
Connect to using your XNAT Session URL and XNAT User ID
Then enter XNAT password to allow access
session = xnat.connect('https://xnat-dev.pm.jh.edu', user = 'bmalik2')
session - Individual XNAT Session (URL)projects - All Accessible Projects dataset - Particular Study in XNAT with various patientsView all accessible projects in XNAT
2 Projects shown here: "Daisy" and "kaggle"
# All accessible Projects
session.projects
<XNATListing {(IRB01234567, Daisy): <ProjectData Daisy (IRB01234567)>, (kaggle, RSNA-MICCAI Brain Tumor Radiogenomic Classification): <ProjectData RSNA-MICCAI Brain Tumor Radiogenomic Classification (kaggle)>}>
Example Dataset used: RSNA-MICCAI Brain Tumor Radiogenomic Classification
session.projects["kaggle"]
<ProjectData RSNA-MICCAI Brain Tumor Radiogenomic Classification (kaggle)>
Number of all Patients within Dataset:
kaggle_project = session.projects["kaggle"]
print("Number of subjects in Dataset: ", len(kaggle_project.subjects))
Number of subjects in Dataset: 672
Can Access list of all Patient Numbers/Labels in respective Project
This allows the user to explore the study with its various patients and imaging series, aid their research analysis by knowing all the patients and number of patients in the study. This list can also be used to modify inclusion criteria.
session.projects["kaggle"].subjects.values() - All Patients in XNAT Study "kaggle"subject.label - Patient IDfor subject in session.projects["kaggle"].subjects.values():
print(subject.label)
00822 00825 00111 00169 00113 00112 00001 00393 00116 00829 00826 00170 00053 00833 00422 00117 00997 00013 00120 00428 00171 00122 00121 00054 00123 00434 01006 00128 00124 00172 00015 00130 00176 00438 00056 00132 00447 00027 00000 00450 00133 00134 00177 00136 00458 00002 00037 00058 00460 00137 00178 00139 00138 00183 00140 00003 00047 00142 00462 00059 00184 00143 00144 00185 00463 00079 00146 00060 00005 00147 00149 00148 00150 00186 00151 00006 00187 00467 00080 00061 00154 00008 00188 00155 00474 00082 00191 00062 00156 00009 00192 00193 00011 00063 00091 00114 00489 00157 00119 00194 00125 00129 00158 00135 00492 00145 00195 00064 00159 00012 00196 00153 00160 00197 00503 00161 00066 00162 00014 00199 00165 00163 00068 00201 00017 00166 00521 00174 00203 00167 00018 00070 00181 00182 00204 00535 00206 00019 00231 00190 00071 00209 00233 00553 00200 00020 00234 00210 00072 00208 00211 00235 00021 00560 00573 00213 00585 00236 00212 00074 00214 00592 00229 00022 00077 00216 00237 00595 00024 00217 00252 00238 00025 00078 00218 00256 00239 00264 00026 00219 00603 00240 00081 00287 00307 00220 00241 00028 00323 00644 00221 00084 00242 00243 00333 00222 00030 00245 00085 00227 00335 00647 00228 00031 00246 00337 00230 00247 00087 00032 00662 00286 00355 00249 00088 00372 00288 00250 00033 00381 00671 00251 00289 00035 00089 00384 00290 00291 00253 00339 00681 00036 00254 00294 00258 00340 00293 00090 00043 00259 00341 00699 00296 00094 00260 00343 00297 00044 00298 00344 00261 00702 00262 00095 00045 00299 00346 00263 00096 00347 00046 00266 00300 00712 00349 00348 00301 00267 00097 00350 00048 00269 00303 00270 00098 00351 00719 00304 00271 00049 00352 00100 00305 00721 00099 00102 00104 00052 00273 00105 00353 00106 00107 00306 00400 00275 00274 00108 00356 00109 00308 00749 00110 00401 00359 00280 00456 00402 00309 00457 00281 00459 00403 00360 00282 00310 00762 00364 00404 00405 00311 00366 00283 00464 00312 00367 00406 00769 00284 00369 00313 00285 00407 00466 00370 00821 00314 00779 00408 00574 00316 00371 00575 00317 00409 00468 00520 00576 00373 00410 00318 00577 00578 00376 00523 00469 00377 00412 00320 00579 00581 00378 00321 00413 00470 00583 00582 00524 00584 00322 00379 00414 00586 00380 00324 00525 00587 00416 00472 00325 00588 00382 00589 00417 00383 00327 00590 00418 00526 00477 00591 00386 00328 00419 00593 00329 00387 00388 00594 00421 00528 00478 00596 00331 00423 00389 00597 00332 00529 00390 00425 00479 00598 00334 00391 00426 00530 00429 00599 00336 00338 00532 00392 00480 00430 00431 00395 00601 00533 00481 00397 00623 00432 00399 00602 00433 00537 00436 00483 00624 00687 00441 00440 00604 00485 00538 00442 00443 00625 00688 00444 00605 00488 00539 00445 00626 00446 00690 00449 00606 00540 00491 00451 00607 00628 00452 00691 00454 00542 00493 00455 00608 00630 00692 00543 00750 00494 00610 00631 00544 00693 00611 00495 00751 00636 00694 00545 00638 00496 00753 00612 00498 00697 00547 00756 00613 00639 00548 00499 00698 00757 00640 00549 00615 00703 00500 00758 00616 00641 00550 00501 00759 00704 00642 00551 00618 00502 00645 00760 00552 00705 00619 00764 00706 00646 00504 00554 00765 00620 00505 00707 00649 00767 00555 00708 00506 00621 00650 00507 00768 00622 00807 00556 00808 00809 00709 00810 00651 00811 00557 00814 00816 00714 00772 00818 00510 00819 00820 00652 00823 00824 00828 00830 00715 00773 00511 00558 00559 00834 00654 00836 00716 00774 00837 00512 00561 00655 00838 00775 00718 00563 00839 00513 00656 00840 00564 00998 00777 00999 00514 00723 00657 01000 00780 00778 00565 00781 00567 01001 00516 00782 01003 01002 00784 00568 00658 00724 00788 00787 00789 00569 01004 00791 00570 01005 00517 00571 00793 00792 00794 00795 00572 01007 00796 00725 00659 00797 00799 01008 00518 00801 01009 00800 00802 00727 00803 00804 00661 00805 01010 00806 00519 00663 00728 00729 00667 00668 00730 00731 00674 00732 00675 00733 00734 00676 00735 00677 00736 00679 00737 00680 00682 00739 00683 00740 00684 00742 00685 00744 00686 00746 00747
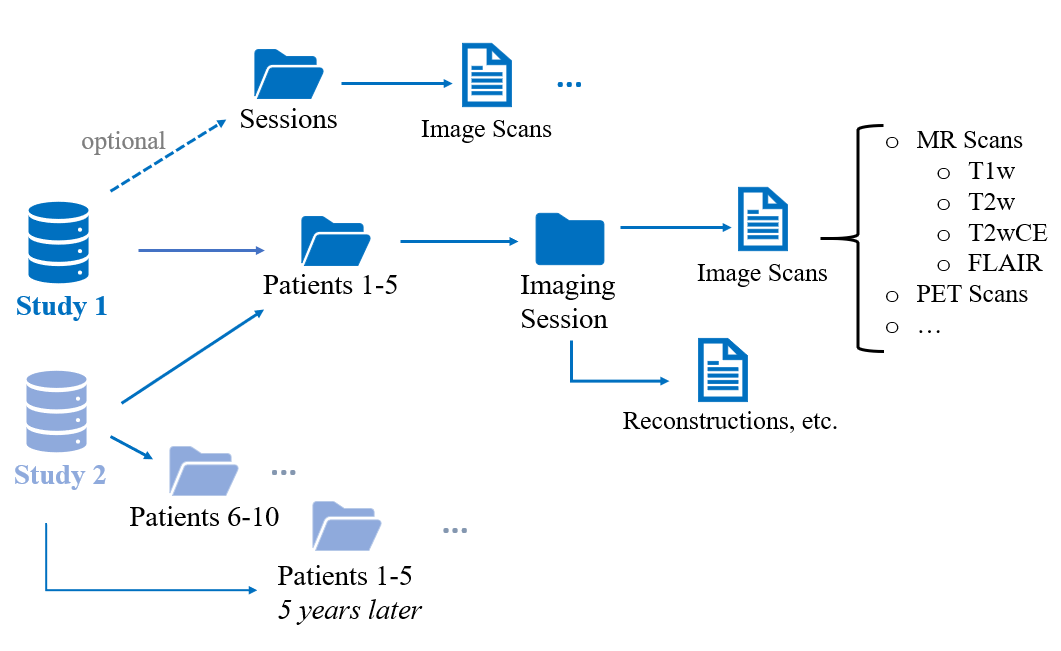
XNAT has specific terminology to its Data Organization and understanding aides in research analysis of the study. The Diagram below explains the organization format.
Dataset > Patient > MR Session > Patient Metadata + Scans
Scans > T2w, T1w, FLAIR, T1wCE

Create File object for particular Subject Scan
File Object - Allows us to use, access and manipulate the particular file
ex: T2w Scan for Patient "00822"
Parameters Include:
Subjects - Individual Patient IDExperiments - Same as Subjects in this exampleScans - Various imaging series available for PatientResources - Includes DICOM and Metadata for particular imaging series. "DICOM" indicates imaging series filefile_object_T2w = session.projects["kaggle"].subjects['00822'].experiments['00822'].scans['T2w'].resources['DICOM'].files[0]
It is crucial to visualize DICOM Data to understand the study and patient imaging series -- which allows researchers to better build their analysis. Instead of managing various files, XNAT allows one to easily view the Patient Imaging series directly in Jupyter Notebook.
Using Pydicom, we read the File Object (T2w image) and display it using matplotlib.pyplot
ds = pydicom.dcmread(file_object_T2w.open())
plt.imshow(ds.pixel_array)
<matplotlib.image.AxesImage at 0x7fc74ad4e2b0>
Accessing DICOM Metadata:
By Accesing the raw DICOM Header Metadata, researchers can better understand the study and modify inclusion/exclusion criteria. Via XNAT, the user does not have to manage metadata and can instead view it easily using the File Object
# DICOM Header Metadata
dicom_metadata = ds
dicom_metadata
Dataset.file_meta ------------------------------- (0002, 0000) File Meta Information Group Length UL: 206 (0002, 0001) File Meta Information Version OB: b'\x00\x01' (0002, 0002) Media Storage SOP Class UID UI: MR Image Storage (0002, 0003) Media Storage SOP Instance UID UI: 1.2.826.0.1.3680043.8.498.12617885361504107518113193736037918908 (0002, 0010) Transfer Syntax UID UI: Explicit VR Little Endian (0002, 0012) Implementation Class UID UI: 1.2.40.0.13.1.1 (0002, 0013) Implementation Version Name SH: 'dcm4che-2.0' (0002, 0016) Source Application Entity Title AE: 'YOURAE' ------------------------------------------------- (0008, 0005) Specific Character Set CS: 'ISO_IR 100' (0008, 0008) Image Type CS: ['DERIVED', 'SECONDARY'] (0008, 0016) SOP Class UID UI: MR Image Storage (0008, 0018) SOP Instance UID UI: 1.2.826.0.1.3680043.8.498.12617885361504107518113193736037918908 (0008, 0050) Accession Number SH: '00822' (0008, 0060) Modality CS: 'MR' (0008, 103e) Series Description LO: 'T2w' (0010, 0010) Patient's Name PN: '00822' (0010, 0020) Patient ID LO: '00822' (0018, 0023) MR Acquisition Type CS: '2D' (0018, 0050) Slice Thickness DS: '5.0' (0018, 0081) Echo Time DS: None (0018, 0083) Number of Averages DS: '1.0' (0018, 0084) Imaging Frequency DS: '123.245481' (0018, 0085) Imaged Nucleus SH: '1H' (0018, 0086) Echo Number(s) IS: '1' (0018, 0087) Magnetic Field Strength DS: '3.0' (0018, 0088) Spacing Between Slices DS: '1.0' (0018, 0089) Number of Phase Encoding Steps IS: '396' (0018, 0091) Echo Train Length IS: '18' (0018, 0093) Percent Sampling DS: '75.0' (0018, 0094) Percent Phase Field of View DS: '100.0' (0018, 0095) Pixel Bandwidth DS: '220.0' (0018, 1310) Acquisition Matrix US: [0, 512, 384, 0] (0018, 1312) In-plane Phase Encoding Direction CS: 'ROW' (0018, 1314) Flip Angle DS: '120.0' (0018, 1316) SAR DS: '0.42682373938699' (0018, 1318) dB/dt DS: '0.0' (0018, 5100) Patient Position CS: 'HFS' (0020, 000d) Study Instance UID UI: 1.2.826.0.1.3680043.8.498.23345897988800591738697565218585968650 (0020, 000e) Series Instance UID UI: 1.2.826.0.1.3680043.8.498.80644139033959530024721674338065709767 (0020, 0011) Series Number IS: '8' (0020, 0013) Instance Number IS: '8' (0020, 0032) Image Position (Patient) DS: [-86.014913593922, -95.009867179948, -17.263729716536] (0020, 0037) Image Orientation (Patient) DS: [0.9874086982475, 0.05804817390263, -0.1471545858332, -0.0973382762022, 0.95623515880841, -0.2759340157441] (0020, 1040) Position Reference Indicator LO: '' (0020, 1041) Slice Location DS: '-54.370920993433' (0028, 0002) Samples per Pixel US: 1 (0028, 0004) Photometric Interpretation CS: 'MONOCHROME2' (0028, 0010) Rows US: 512 (0028, 0011) Columns US: 512 (0028, 0030) Pixel Spacing DS: [0.390625, 0.390625] (0028, 0100) Bits Allocated US: 16 (0028, 0101) Bits Stored US: 16 (0028, 0102) High Bit US: 15 (0028, 0103) Pixel Representation US: 0 (0028, 1050) Window Center DS: '742.0' (0028, 1051) Window Width DS: '1506.0' (0028, 1052) Rescale Intercept DS: '0.0' (0028, 1053) Rescale Slope DS: '1.0' (0028, 1054) Rescale Type LO: 'US' (6781, 1000) Private tag data UN: b'Project:kaggle' (7fe0, 0010) Pixel Data OW: Array of 524288 elements
Users can locally download Patient Imaging Series if they need local access to the file.
download_dirrequires path to file where Patient Series should be downloaded to.
# Locate Patient scan, then download
scan_to_download = session.projects["kaggle"].subjects['XNAT_S00007'].experiments['XNAT_E00006'].scans['T2w']
scan_to_download.download_dir('./')